 PDF(1304 KB)
PDF(1304 KB)


 PDF(1304 KB)
PDF(1304 KB)
 PDF(1304 KB)
PDF(1304 KB)
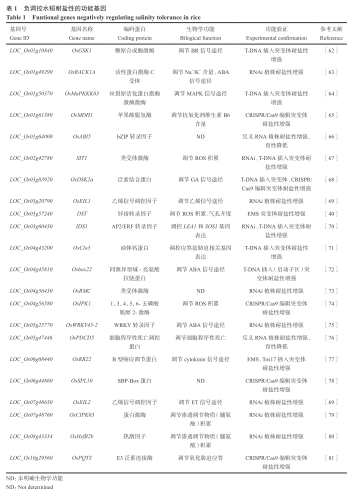
水稻耐盐分子机制与分子改良
Molecular Mechanisms and Improvement of Salinity Tolerance in Rice
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |